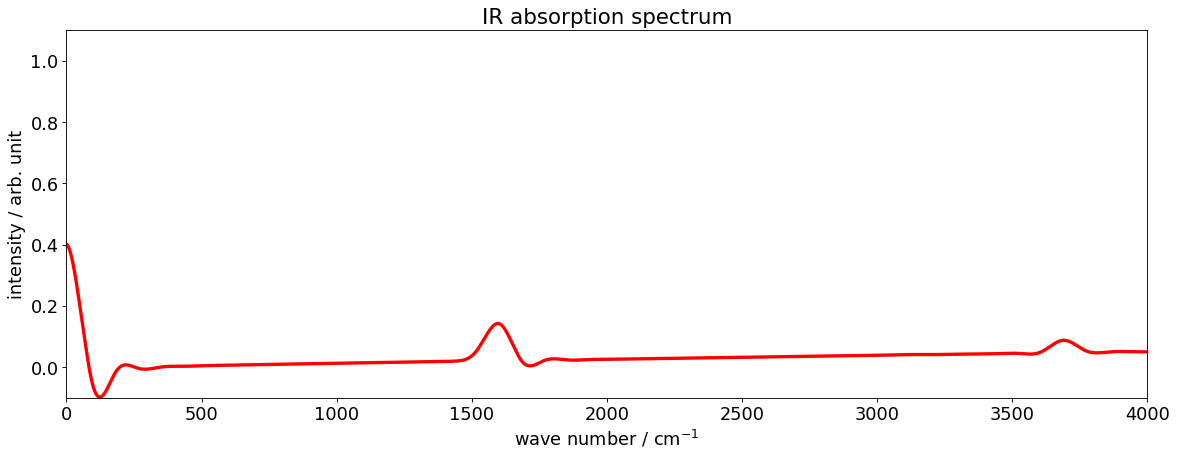
Example 4: Plot IR spectrum of H2O molecule
Documentation is here
The IR absorption spectrum is obtained by the Fourier transformation procedure
\[\begin{align}
I_r(\omega)
&\simeq\frac{\pi\omega}{3c\epsilon_0\hbar^2}
\frac{Re}{\pi}\int_0^T e^{i(\hbar\omega+E_0)t} g(t)a_r(t) dt
\end{align}\]
where \(a_r(t)=\langle\Psi_\mu(0)|\Psi_\mu(t)\rangle\) is an autocorrelation function, \(E_0\) can be regarded as the vibrational ground state energy and \(g(t)\) is the window function, defaults to \(g(t)=\cos^2\left(\frac{\pi t}{2T}\right)\)
1. Check autocorrelation file
[1]:
!ls h2o_polynomial_prop/autocorr*
h2o_polynomial_prop/autocorr.dat
2. Fourier transform and plot
By the so-called t/2-trick, the correlation time is twice the propagation time.
[2]:
import pytdscf
E_0 = 0.5675130 # estimated zero point energy in example 1
time, autocorr = pytdscf.spectra.load_autocorr(
"h2o_polynomial_prop/autocorr.dat"
)
pytdscf.spectra.plot_autocorr(time, autocorr, gui=False)
freq, intensity = pytdscf.spectra.ifft_autocorr(time, autocorr, E_shift=E_0)
pytdscf.spectra.plot_spectrum(
freq,
intensity,
lower_bound=0,
upper_bound=4000,
show_in_eV=False,
gui=False,
)
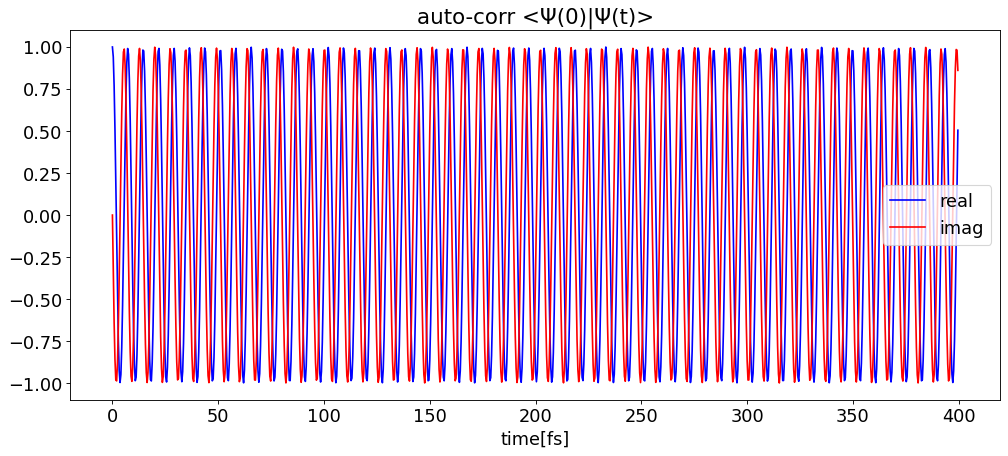
delta_t: 0.200[fs], max_freq: 166782.0[cm-1]